Even if two datasets are derived from the same pool of observations, it could still be the case that there are unaccounted for differences between the two datasets that are not apparent to a human observer. Any such discrepancies could change the way we think about the data, and could, for example, justify building two separate models, or suggest the existence of inconsistencies in the way the datasets were generated, undermining the validity of any inferences drawn from the datasets. Below, I’ll show how we can use one of my algorithms to measure the internal consistency of a single dataset, and the consistency between two datasets.
Measuring Internal Consistency
In a previous article, I mentioned that I had written an algorithm that can quickly generate non-mutually exclusive categories on a dataset. Like nearly all of my algorithms, this “graph categorization algorithm” generates a measure of distinction  , that tells us how different two data points need to be in order to justify distinguishing between them in the context of the dataset. Specifically, if vectors
, that tells us how different two data points need to be in order to justify distinguishing between them in the context of the dataset. Specifically, if vectors  and
and 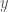 are both in some dataset, then we distinguish between
are both in some dataset, then we distinguish between  and
and  only if
only if  . That is, if two vectors are within
. That is, if two vectors are within  of each other, then we treat them as equivalent, whereas if the norm of their difference exceeds
of each other, then we treat them as equivalent, whereas if the norm of their difference exceeds  , then we distinguish between them.
, then we distinguish between them.
The structure of the dataset will of course affect the value of  . Generally speaking, if the data is spread out, then
. Generally speaking, if the data is spread out, then  will be large, and if the data is concentrated, then
will be large, and if the data is concentrated, then  will be small. As a result, when we add new data to a dataset, we will almost certainly change the value of
will be small. As a result, when we add new data to a dataset, we will almost certainly change the value of  . However, if we add new data that is drawn from the same underlying dataset, then the value of
. However, if we add new data that is drawn from the same underlying dataset, then the value of  shouldn’t change much. That is, if the original dataset is sufficiently large, then we’re not learning anything from the new data – we’re just including more examples of the same type of data. As a result, we can use
shouldn’t change much. That is, if the original dataset is sufficiently large, then we’re not learning anything from the new data – we’re just including more examples of the same type of data. As a result, we can use  as a measure of how much the inclusion of new data changes the structure of a dataset, by evaluating
as a measure of how much the inclusion of new data changes the structure of a dataset, by evaluating  before and after the inclusion of the new data.
before and after the inclusion of the new data.
Let’s begin by incrementally adding new data to a dataset, and measuring the value of  at each iteration. The dataset will in this case consist of 10-dimensional vectors of random numbers generated by Octave. The expectation is that the value of
at each iteration. The dataset will in this case consist of 10-dimensional vectors of random numbers generated by Octave. The expectation is that the value of  should stabilize once the dataset is sufficiently large, since once we have enough data, the appropriate level of distinction should become clear, and roughly invariant with respect to new data being added. Stated differently, assuming my algorithms work, there should be a single level of distinction for a dataset, that shouldn’t change as we add new data, assuming the new data is of the same type as the existing data.
should stabilize once the dataset is sufficiently large, since once we have enough data, the appropriate level of distinction should become clear, and roughly invariant with respect to new data being added. Stated differently, assuming my algorithms work, there should be a single level of distinction for a dataset, that shouldn’t change as we add new data, assuming the new data is of the same type as the existing data.
We can accomplish this with the following Octave code:
N = 10;
for i = 1 : 500
data_matrix = rand(i,N);
[final_graph_matrix final_delta] = generate_dataset_graph(data_matrix, N);
data_vector(i) = final_delta;
final_delta
endfor
figure, plot(1:500,data_vector)
This will add one new vector at a time to the dataset, categorize the dataset using the “graph categorization algorithm”, print the resultant value of  , and store it in a vector that is later displayed as a graph. Below is the graph generated by the code above, which will vary somewhat each time you run the code, since the dataset is generated randomly. Note that even though the graph categorization algorithm is fast, and has a polynomial runtime, the code above involves calling the algorithm
, and store it in a vector that is later displayed as a graph. Below is the graph generated by the code above, which will vary somewhat each time you run the code, since the dataset is generated randomly. Note that even though the graph categorization algorithm is fast, and has a polynomial runtime, the code above involves calling the algorithm  times, so it’s going to take a little while to run. I’ve named the graph generated by plotting
times, so it’s going to take a little while to run. I’ve named the graph generated by plotting  as a function of the size of the dataset the consistency curve for the dataset. The average over the consistency curve is in this case
as a function of the size of the dataset the consistency curve for the dataset. The average over the consistency curve is in this case  , and the standard deviation is
, and the standard deviation is  .
.
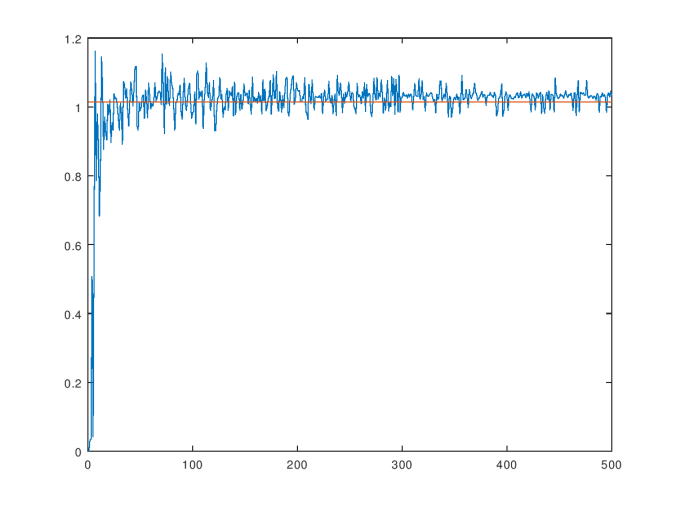
The consistency curve for the dataset of random vectors.
As you can see, the value of  stabilizes near the average, which is shown as the orange line in the graph above. This result is consistent with the hypothesis that there should be a single objective value of
stabilizes near the average, which is shown as the orange line in the graph above. This result is consistent with the hypothesis that there should be a single objective value of  that is intrinsic to the dataset in the abstract, and not dependent upon the number of observations. Obviously, without a sufficiently large number of observations, you can’t determine this value. But nonetheless, at the risk of being overly philosophical, there really is an underlying process that generates every dataset. Therefore, the correct minimum difference that warrants distinction in the context of a particular dataset is a function of that process, not the observed dataset. The observed dataset gives us a window into that process, and allows us to generate an approximation of the true, underlying in-context difference that warrants distinction between observations generated by that process.
that is intrinsic to the dataset in the abstract, and not dependent upon the number of observations. Obviously, without a sufficiently large number of observations, you can’t determine this value. But nonetheless, at the risk of being overly philosophical, there really is an underlying process that generates every dataset. Therefore, the correct minimum difference that warrants distinction in the context of a particular dataset is a function of that process, not the observed dataset. The observed dataset gives us a window into that process, and allows us to generate an approximation of the true, underlying in-context difference that warrants distinction between observations generated by that process.
The idea that there is a single, in-context measure of distinction for a dataset is further supported by the observation that replacing the graph categorization algorithm, with my original categorization algorithm (“optimize_categories_N”), in the code above produces very similar values for  . This is actually remarkable, because these two algorithms make use of different processes, and generate different outputs: the graph categorization algorithm generates non-mutually exclusive categories, whereas the original categorization algorithm generates mutually exclusive categories. Nonetheless, they generate approximately the same values of
. This is actually remarkable, because these two algorithms make use of different processes, and generate different outputs: the graph categorization algorithm generates non-mutually exclusive categories, whereas the original categorization algorithm generates mutually exclusive categories. Nonetheless, they generate approximately the same values of  , which supports the notion that just like a dataset has a “true” mean, and standard deviation, there is also a true minimum difference that warrants distinction – i.e., a true value of
, which supports the notion that just like a dataset has a “true” mean, and standard deviation, there is also a true minimum difference that warrants distinction – i.e., a true value of  .
.
It also supports the idea that my method of selecting the level of distinction that generates the greatest change in the entropy of the object in question is the correct way to find this value of  , since this is the method that both algorithms have in common, despite the fact that they produce different outputs using that value of
, since this is the method that both algorithms have in common, despite the fact that they produce different outputs using that value of  .
.
Now let’s repeat the same process using the Wine Dataset, which is courtesy of the UCI Machine Learning Repository. The same hypothesis should hold, which is that the value of  should stabilize around the “correct”, in-context level of distinction for the dataset as we add more data. This is in fact exactly what happens, as you can see in the chart below. The average value over the consistency curve is in this case
should stabilize around the “correct”, in-context level of distinction for the dataset as we add more data. This is in fact exactly what happens, as you can see in the chart below. The average value over the consistency curve is in this case 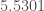 , and the standard deviation is
, and the standard deviation is  .
.

The consistency curve for the Wine Dataset.
The volatility of the consistency curve should decline as a function of the number of observations, and if it doesn’t, then this implies that new observations carry new information about the structure of the dataset. Since this could of course be the case, not all datasets will produce consistency curves that stabilize, and so we can use this curve as a measure of the internal consistency of a dataset. That is, the greater the volatility of this curve, the more “shocks” there are to the structure of the dataset from new observations. If this persists, then there might not be a single process at work generating the data, which means that what we’re observing might actually be transitory in nature, and not a stable process that we can observe and predict. Alternatively, it could be the case that the underlying process has a high complexity. That is, if the underlying process has a high Kolmogorov complexity, then it will generate a large number of novel observations, each of which will shock the dataset. Finally, the dataset could also contain interjections of noise, which will also shock the dataset if the noise is novel every time, which is plausible, since noise is presumably the product of a Kolmogorov-random process that generates novel observations.

The consistency curve for the Ionosphere Dataset.
Above is the consistency curve for the Ionosphere Dataset, which is also courtesy of the UCI Machine Learning Repository. The average over the curve is  , and the standard deviation is
, and the standard deviation is 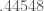 . As you can see, it’s highly volatile, suggesting that new observations significantly change the structure of the dataset. Interestingly, this dataset produces a large percentage of “rejected” inputs when my prediction algorithm is applied to it. A rejected input indicates that the prediction algorithm believes that the input is beyond the scope of the training dataset. If a dataset is not internally consistent, then randomly selected observations are more likely to be novel observations, and therefore, outside the scope of the training dataset. Therefore, we would expect a large percentage of rejections for a dataset that is not internally consistent, which is exactly what happens in the case of the Ionosphere Dataset.
. As you can see, it’s highly volatile, suggesting that new observations significantly change the structure of the dataset. Interestingly, this dataset produces a large percentage of “rejected” inputs when my prediction algorithm is applied to it. A rejected input indicates that the prediction algorithm believes that the input is beyond the scope of the training dataset. If a dataset is not internally consistent, then randomly selected observations are more likely to be novel observations, and therefore, outside the scope of the training dataset. Therefore, we would expect a large percentage of rejections for a dataset that is not internally consistent, which is exactly what happens in the case of the Ionosphere Dataset.
It’s tempting to think that a countably infinite number of observations (which is obviously not physically possible), would allow us to discern the “true” level of distinction using this process, but I’m not sure that’s correct. I haven’t worked through the mathematics carefully, yet, but even a superficial analysis implies that the notion of entropy has to change when you have a countable set, since Shannon’s equation does not work with a countable set. Specifically, you can’t have a uniform distribution on a countable set using ordinary probabilities, and therefore, you need a new measure of entropy if you’re going to make use of countable sets. But, as far as we know, observation is finite, so this question is academic, at least for now.
Measuring Consistency Between Datasets
If we have two datasets of observations that were ostensibly generated using the same procedures, and sampled from the same source, then the value of  shouldn’t change much when we combine the datasets. If
shouldn’t change much when we combine the datasets. If  does change significantly, then either the two datasets are incomplete on their own, or, there’s some underlying difference between them that’s unaccounted for.
does change significantly, then either the two datasets are incomplete on their own, or, there’s some underlying difference between them that’s unaccounted for.
There are probably a number of reasonable ways to go about testing the consistency between two datasets using  , but the method I’ve decided to use is to generate three consistency curves: one for the first dataset, one for the second dataset, and one for the combined dataset. Then, we can measure both the average value and standard deviation of each consistency curve. When examining the consistency curve for the combined dataset, if it turns out that the average value changes significantly, or the standard deviation increases significantly, in each case as compared to the two individual curves, then it suggests that combining the datasets significantly disturbed the structure of the individual datasets. This in turn suggests that the two datasets are in fact distinct. In contrast, if the average value of
, but the method I’ve decided to use is to generate three consistency curves: one for the first dataset, one for the second dataset, and one for the combined dataset. Then, we can measure both the average value and standard deviation of each consistency curve. When examining the consistency curve for the combined dataset, if it turns out that the average value changes significantly, or the standard deviation increases significantly, in each case as compared to the two individual curves, then it suggests that combining the datasets significantly disturbed the structure of the individual datasets. This in turn suggests that the two datasets are in fact distinct. In contrast, if the average value of  is not significantly changed, and the standard deviation is unchanged or decreases, then it suggests that the two datasets are consistent.
is not significantly changed, and the standard deviation is unchanged or decreases, then it suggests that the two datasets are consistent.
If the two datasets are both reasonably large, and their individual consistency curves stabilize, then if the combined consistency curve is drastically more volatile, we can be more confident that the datasets are not incomplete, but that instead, there is some bona fide difference between them. If they’re both sampled from the same source, then there must be some difference in the sampling process that explains the resultant differences between the datasets. As a result, we can also use this process to identify inconsistencies in sampling methods used to gather data, as well as distinguish between datasets that are superficially similar, but nonetheless have some subtle differences that might not be apparent to a human observer.
We’ll apply this process by combining the Parkinsons Dataset, which is again courtesy of the UCI Machine Learning Repository, and the Wine Dataset. The Parkinsons Dataset is 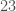 dimensions, and the Wine Dataset is
dimensions, and the Wine Dataset is 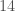 dimensions. As a result, we can’t combine them without reducing the dimension of the Parkinsons Dataset to
dimensions. As a result, we can’t combine them without reducing the dimension of the Parkinsons Dataset to 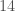 . Reducing the dimension of the Parkinsons Dataset will obviously affect the dataset, but we’re using it for a very limited purpose, which is to demonstrate that when two datasets that are clearly not consistent with each other are combined, the consistency curve will be drastically impacted.
. Reducing the dimension of the Parkinsons Dataset will obviously affect the dataset, but we’re using it for a very limited purpose, which is to demonstrate that when two datasets that are clearly not consistent with each other are combined, the consistency curve will be drastically impacted.

The consistency curve for the Parkinsons Dataset, limited to  .
.
Above is the consistency curve for the Parkinsons Dataset, limited to 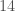 dimensions of data. The average over the curve is
dimensions of data. The average over the curve is  , and the standard deviation is
, and the standard deviation is  . Though there are some shocks, it trends reasonably close to the average, suggesting that the dataset is reasonably internally consistent, even when limited to
. Though there are some shocks, it trends reasonably close to the average, suggesting that the dataset is reasonably internally consistent, even when limited to 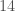 dimensions. Below is the consistency curve for the Parkinsons Dataset using the full
dimensions. Below is the consistency curve for the Parkinsons Dataset using the full 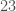 dimensions of data. The average over the curve below is
dimensions of data. The average over the curve below is  , and the standard deviation is
, and the standard deviation is  . The inclusion of the additional
. The inclusion of the additional 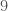 dimensions obviously affected the consistency curve significantly, but this was expected.
dimensions obviously affected the consistency curve significantly, but this was expected.

The consistency curve for the Parkinsons Dataset, using all 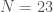 dimensions.
dimensions.
I combined the two datasets into a single dataset with the rows from the Wine Dataset first, and the rows from the Parkinsons Dataset afterward, without changing the order of the rows in either dataset. I then ran the same process on the combined dataset, which generated the consistency curve below. The Wine Dataset contains 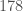 rows, and the Parkinsons Dataset contains
rows, and the Parkinsons Dataset contains 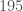 rows, and you can clearly see that the consistency curve breaches the average after roughly
rows, and you can clearly see that the consistency curve breaches the average after roughly  observations, suggesting that the inclusion of the Parkinsons Dataset drastically altered the dataset, which is consistent with our hypothesis. The average over the curve is
observations, suggesting that the inclusion of the Parkinsons Dataset drastically altered the dataset, which is consistent with our hypothesis. The average over the curve is  , and the standard deviation is
, and the standard deviation is  .
.

The consistency curve for the combined dataset.
Deciding whether two datasets are distinct is a binary question, but the purpose of this process is to provide data that informs a decision either way, rather than an objective threshold for distinction. In this case, the results are rather obvious. Nonetheless, the decision to distinguish will depend upon what you’re doing with the data. That is, even if there’s a significant degree of inconsistency between two datasets, it might not matter for certain purposes, which means that we can’t set an objective point at which distinction is necessary, without also taking into account the purpose for which the datasets are being used.
This process of comparing consistency curves could be a powerful tool for statisticians looking to identify errors and inconsistencies in their sampling procedures, and for data scientists deciding whether to build separate models for ostensibly similar datasets. Though it’s not an area of interest for me, I suspect this methodology could also be used to facilitate forgery detection, and DNA analysis, since this process would uncover discrepancies in superficially similar datasets, which could be generated by examining real-world objects.
Applications to Data Compression
If we have an extremely large dataset, then we probably don’t want to use the entire dataset as a training dataset, since this will require a significant amount of time, and, at least when using my software, generate a large model (i.e., the data structure that models the dataset will be large). A large model will slow down predictions, so for both reasons, we should use as little data as possible in training the learning algorithm.
In order to compress a dataset, we could randomly select data from the dataset until the consistency curve stabilizes. Once the consistency curve stabilizes, we can form the reasonable expectation that there shouldn’t be any further shocks to the sampled dataset from new observations, and therefore, any new information from the dataset will be of only marginal importance. Of course, this could be wrong, and there could be some corner of the dataset that is significant, that we just happened to miss in our sample. But nonetheless, as a practical matter, this should work just fine, and if at some future point our model starts to generate a significant number of errors, then we can retrain it.
Changes in Entropy; Physics
For those that are interested in further reading, in a previous article, I discussed how the entropy of a dataset changes as we add new data. The results are similar to the results presented above, and actually form the basis of my prediction algorithm. For those interested in making sense of the Kolmogorov complexity of a physical process, you can see my paper on the applications of information theory to physics, though it is quite long.





